Jaro-Winkler
Saturday, June 21, 2025
Jaro-Winkler distance is a measure of string similarity and edit distance between two sequences:
The higher the Jaro–Winkler distance for two strings is, the less similar the strings are. The score is normalized such that 0 means an exact match and 1 means there is no similarity. The original paper actually defined the metric in terms of similarity, so the distance is defined as the inversion of that value (distance = 1 − similarity).
There are actually two different concepts – and RosettaCode tasks – implied by this algorithm:
- Jaro similarity and Jaro distance
- Jaro-Winkler similarity and Jaro-Winkler distance.
Let’s build an implementation of these in Factor!
Jaro Similarity
The base that all of these are built upon is the Jaro similarity. It is
calculated as a score by measuring the number of matches (m) between the
strings, counting the number of transpositions divided by 2 (t), and then returning a
weighted score using the formula using the lengths of each sequence (|s1|
and |s2|):
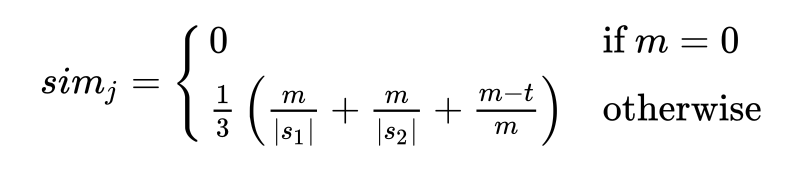
In particular, it considers a matching character to be one that is found in the other string within a match distance away, calculated by the formula:
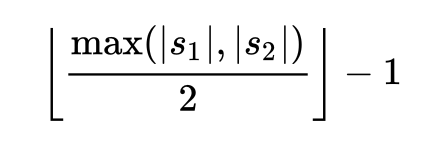
There are multiple ways to go about this, with varying performance, but I decided one longer function was simpler to understand than breaking out the steps into their own words. We use a bit-array to efficiently track which characters have been matched already as we iterate:
:: jaro-similarity ( s1 s2 -- n )
s1 s2 [ length ] bi@ :> ( len1 len2 )
len1 len2 max 2/ 1 [-] :> delta
len2 <bit-array> :> flags
s1 [| ch i |
i delta [-] :> from
i delta + 1 + len2 min :> to
from to [| j |
j flags nth [ f ] [
ch j s2 nth = dup j flags set-nth
] if
] find-integer-from
] filter-index
[ 0 ] [
[ length ] keep s2 flags [ nip ] 2filter [ = not ] 2count
:> ( #matches #transpositions )
#matches len1 /f #matches len2 /f +
#matches #transpositions 2/ - #matches /f + 3 /
] if-empty ;
The Jaro distance is then just a subtraction:
: jaro-distance ( s1 s2 -- n )
jaro-similarity 1.0 swap - ;
I’m curious if anyone else has a simpler implementation – please share!
Jaro-Winkler Similarity
The Jaro-Winkler similarity builds upon this by factoring in the length of
the common prefix (l) times a constant scaling factor (p) that is
usually set to 0.1 in most implementations I’ve seen:
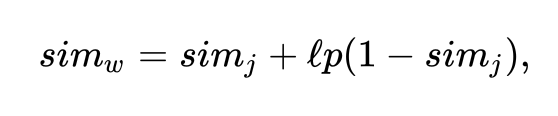
We can implement this by calcuting the Jaro similarity and then computing the common prefix and then generating the result:
:: jaro-winkler-similarity ( s1 s2 -- n )
s1 s2 jaro-similarity :> jaro
s1 s2 min-length 4 min :> len
s1 s2 [ len head-slice ] bi@ [ = ] 2count :> #common
1 jaro - #common 0.1 * * jaro + ;
The Jaro-Winkler distance is again just a subtraction:
: jaro-winkler-distance ( a b -- n )
jaro-winkler-similarity 1.0 swap - ;
Try it out
The Wikipedia article
compares the similarity of FARMVILLE and FAREMVIEL:
IN: scratchpad "FARMVILLE" "FAREMVIEL" jaro-similarity .
0.8842592592592592
We can also see that the algorithm considers the transposition of two close characters to be less of a penalty than the transposition of two characters farther away from each other. It also penalizes additions and substitutions of characters that cannot be expressed as transpositions.
IN: scratchpad "My string" "My tsring" jaro-winkler-similarity .
0.9740740740740741
IN: scratchpad "My string" "My ntrisg" jaro-winkler-similarity .
0.8962962962962963
We can compare the rough performance of Julia using the same algorithm:
julia> using Random
julia> s = randstring(10_000)
julia> t = randstring(10_000)
julia> @time jarowinklerdistance(s, t)
1.492011 seconds (108.32 M allocations: 2.178 GiB, 1.87% gc time)
0.19016926812348256
Note: I’m not a Julia developer, I just play one on
TV. I adapted this
implementation in
Julia, which
originally took over 4.5 seconds. A better developer could probably improve it
quite a bit. In fact, it was pointed out that we are indexing UTF-8 String
in a loop, and should instead collect the Char into a Vector first.
That does indeed make it super fast.
To the implementation in Factor that we built above, which runs quite a bit faster:
IN: scratchpad USE: random.data
IN: scratchpad 10,000 random-string
10,000 random-string
gc [ jaro-winkler-distance ] time .
Running time: 0.259643166 seconds
0.1952856823031448
Thats not bad for a first version that uses safe indexing with unnecessary bounds-checking, generic iteration on integers when usually the indices are fixnum (something I hope to fix someday automatically), and should probably order the input sequences by length for consistency.
If we fix those problems, it gets even faster:
IN: scratchpad USE: random.data
IN: scratchpad 10,000 random-string
10,000 random-string
gc [ jaro-winkler-distance ] time .
Running time: 0.068086625 seconds
0.19297898770334765
This is available in the development version in the math.similarity and the math.distances vocabularies.